The idea of simultaneously analyzing medical diagnosis codes with electronic medical records has been much like knowing valuable treasure is buried under miles of rock but lacking the tools to mine it.
The payoff—connections that may detect disease earlier and identify new research paths—has tantalized engineers and clinicians alike, remaining largely out of reach.
But not for long.

A new tool developed by Vanderbilt engineers, in collaboration with clinical and informatic experts, can unearth novel co-morbidities from routinely collected, anonymized electronic medical records. The team, led by Bennett Landman, associate professor of electrical engineering, computer engineering and computer science, tested the tool for three conditions. In all three—Alzheimer’s Disease, Autism Spectrum Disorder and Optic Neuritis, which can be the first indication of multiple sclerosis—the new tool found lesser-known conditions that may support earlier monitoring or medical intervention. The novel connections also suggest new research opportunities.
“We are excited about the opportunities to discover new risk factors and associations of diseases in the clinical record,” Landman said. “Overall, our goal is to advance engineering and clinical science to improve the understanding and care of patients.”
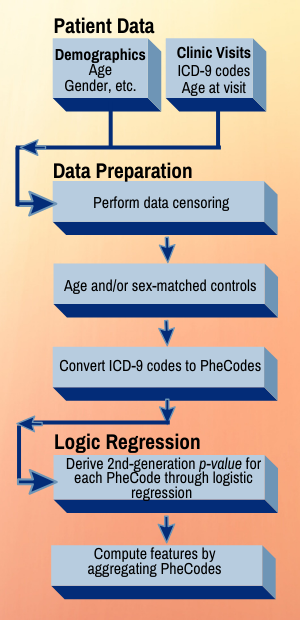
The team’s tool, Phenome-Disease Association Study, or PheDAS, performs association studies and identifies disease comorbidities across time in electronic medical records. It also solves an issue that has vexed researchers for years: how to prioritize apparent correlations for clinical relevance. PheDAS correctly identified well-known associations with each of the three target conditions. But some associations will be so random they are likely to be unrelated or have extremely limited relevance.
A new “Novel Finding Index” guides researchers to significant associations that may be clinically relevant but have not been well-studied in medically literature. The index gives well-known disease associations a low ranking.

3 studied conditions validate tool and Novel Finding Index
In the case of Alzheimer’s, for example, well-known associations identified included psychosis, cerebral degenerations and gait abnormalities and were given a low novelty score. Infections and inflammatory processes across several organ systems were among the novel associations identified in the five years prior to diagnosis and received higher scores.
The three example conditions validated the tool, which researchers can use to investigate other diseases if they have their own approved data sets from the Synthetic Derivative, the database containing anonymized clinical information derived from Vanderbilt’s electronic medical record of 2.2 million people, the Baltimore Longitudinal Study on Aging, or similar data from other institutions. In part because of this study, data from medical imaging is now added to the Synthetic Derivative as well under the umbrella of BioVU, Vanderbilt’s repository of DNA extracted from discarded blood collected during routine clinical testing and linked to de-identified medical records.
The new tool kit, with its machine learning algorithms, creates easier, user-friendly access to a daunting amount of data.
“Our lab primarily focuses on medical imaging, including magnetic resonance imaging and computed tomography,” Landman said. “These new tools will allow us to better interpret imaging findings in the context of a patient’s broader story.”
Plus One, an open-access, peer-reviewed journal, published the findings online in November 2019. Clinical collaborators with interest in the example cases already are digging into specific findings for each condition. The large team includes experts from the departments of behavioral neurosciences, psychiatry and behavioral sciences, ophthalmology and visual sciences, neurology, radiology and radiological sciences, biostatistics and biomedical informatics.
Researchers used de-identified EMRs of patient groups with each of the three conditions and appropriate control groups with comparable demographics but without the disease diagnosis. They mined real-time journal article abstracts from PubMed, a search engine maintained by the U.S. Library of Medicine, because well-known associations likely will have more papers published about them than novel ones, said Shikha Chaganti, Ph.D’19 and a former student in Landman’s Medical-image Analysis and Statistical Interpretation Lab.
Project incorporates PubMed, ICD codes, EMRs, and PheCodes
The algorithms searched for and tallied mentions of associations to each condition from article headlines, abstracts and keywords.
“We found 194,736 articles with search terms related to Alzheimer’s Disease, 45,419 for Autism Spectrum Disorder terms, and 18,894 on Optic Neuritis terms,” said Chaganti, the study’s first author. She is now at Siemens Healthineers as a senior deep learning research scientist.
Roughly 15,000 codes from the International Classification of Diseases (ICD) system were mapped to 1,865 PheCodes, which are combinations of ICD codes for distinct diseases, traits or conditions. PheCodes were pioneered by Joshua Denny, a Vanderbilt professor of biomedical informatics and medicine, and now widely used to interpret billing records. (Denny was appointed CEO of All of Us, the NIH precision health research program, in December 2019.)
Associations are ranked by comparing each PheCode-disease finding to the number of papers found on PubMed that mention both the diagnosis and the associated condition as a proportion of the number of papers published on the disease of interest. By adjusting for well-known associations and those with less likely clinical relevance, the novelty score moves the PubMed proportion from an absolute scale onto a relative scale.
“Our results demonstrate wide utility for identifying new associations in EMR data that have the highest priority among the complex web of correlations and causalities,” the team concluded. “Data scientists and clinicians can work together more effectively to discover novel associations that are both empirically reliable and clinically understudied.”

Contact:
Pamela Coyle (615) 343-5495
Pam.Coyle@Vanderbilt.edu
Twitter @VUEngineering